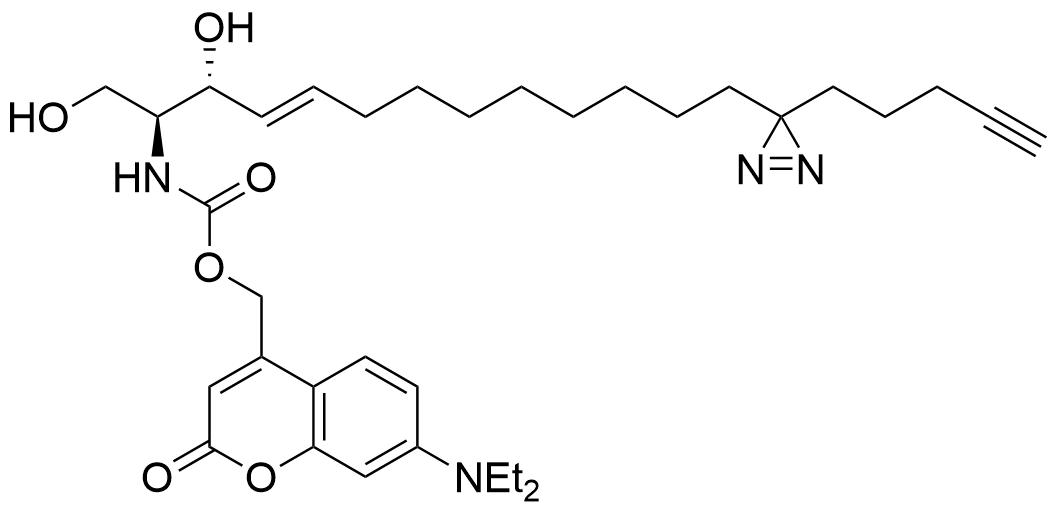
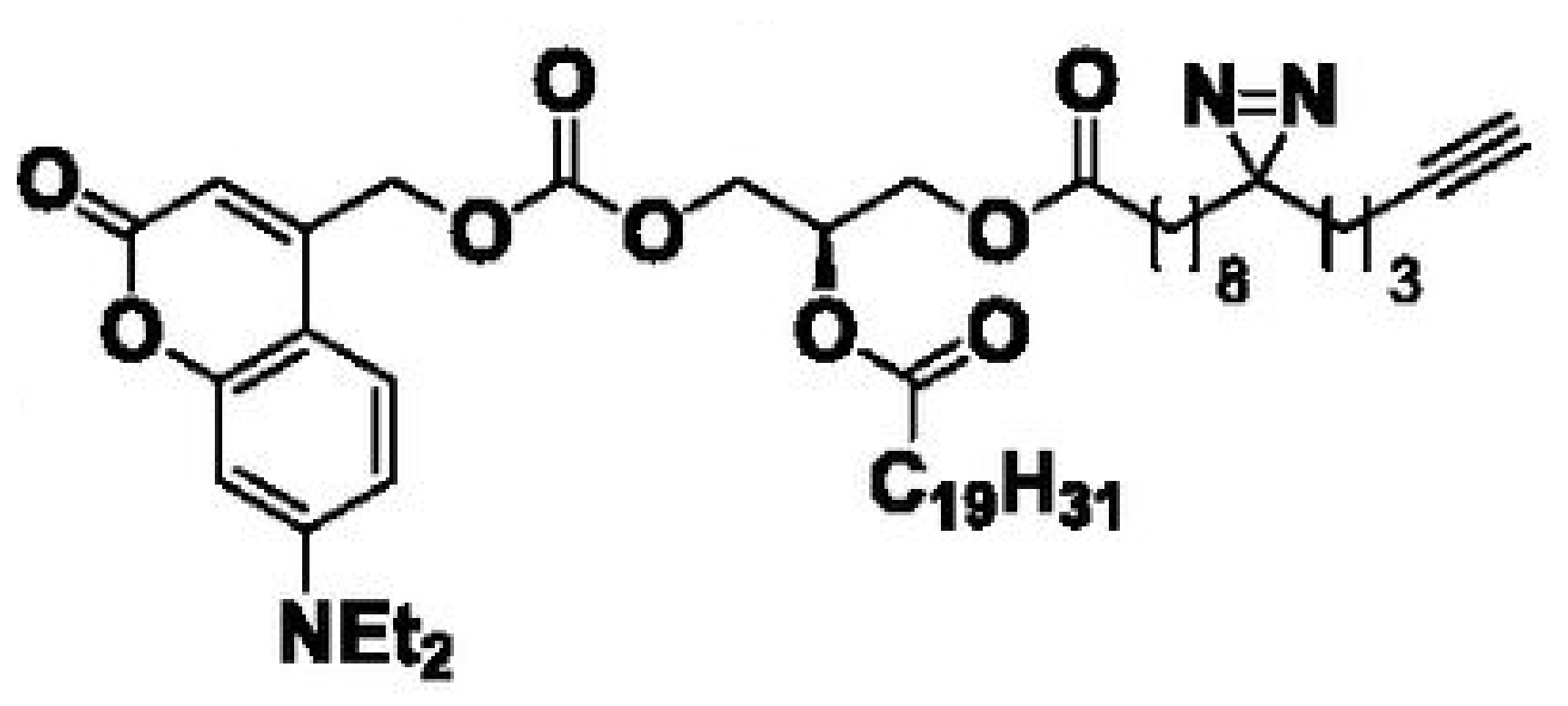
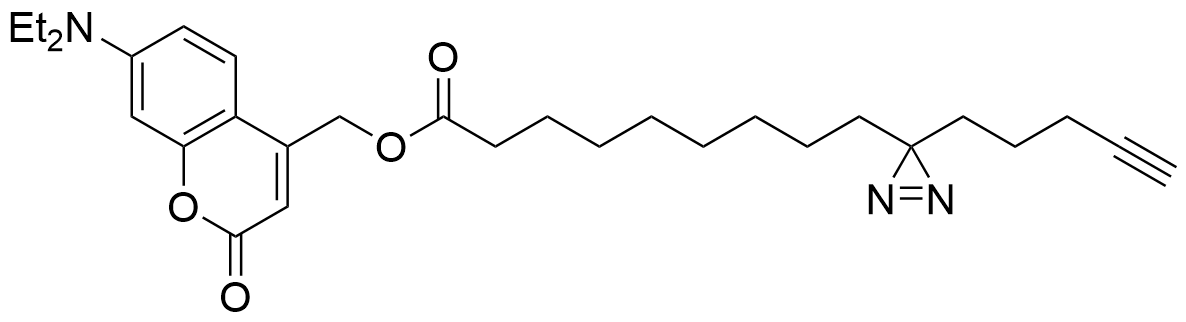
Heat map depicts the average PSM count for two replicates each of UV-irradiated samples treated with trifunctionalized Sph, DAG, and FA. Color denotes the
Höglinger et al. 2017
Trifunctional lipid probes for comprehensive studies of single lipid species in living cells
Journal
PNAS
Abstract
Lipid-mediated signaling events regulate many cellular processes. Investigations of the complex underlying mechanisms are difficult because several different methods need to be used under varying conditions. Here we introduce multifunctional lipid derivatives to study lipid metabolism, lipid−protein interactions, and intracellular lipid localization with a single tool per target lipid. The probes are equipped with two photoreactive groups to allow photoliberation (uncaging) and photo–cross-linking in a sequential manner, as well as a click-handle for subsequent functionalization. We demonstrate the versatility of the design for the signaling lipids sphingosine and diacylglycerol; uncaging of the probe for these two species triggered calcium signaling and intracellular protein translocation events, respectively. We performed proteomic screens to map the lipid-interacting proteome for both lipids. Finally, we visualized a sphingosine transport deficiency in patient-derived Niemann−Pick disease type C fibroblasts by fluorescence as well as correlative light and electron microscopy, pointing toward the diagnostic potential of such tools. We envision that this type of probe will become important for analyzing and ultimately understanding lipid signaling events in a comprehensive manner.
Lipid probes utilized
Cell line analyzed
Species: Homo sapiens
Cell type: Epithelial (immortalized cervical cancer line)
Uncaging & Crosslinking timeline
| Lipid Probe | Uptake time | Uncaging time | Interaction time | Crosslinking time |
|---|---|---|---|---|
| Sph | 5 min | 2.5 min | 0 min | 2.5 min |
| DAG | 15 min | 2.5 min | 0 min | 2.5 min |
| 8-3 FA | 15 min | 2.5 min | 0 min | 2.5 min |
Mass spectrometry quantification method
Peptide spectral counting
Data visualization
Heat map
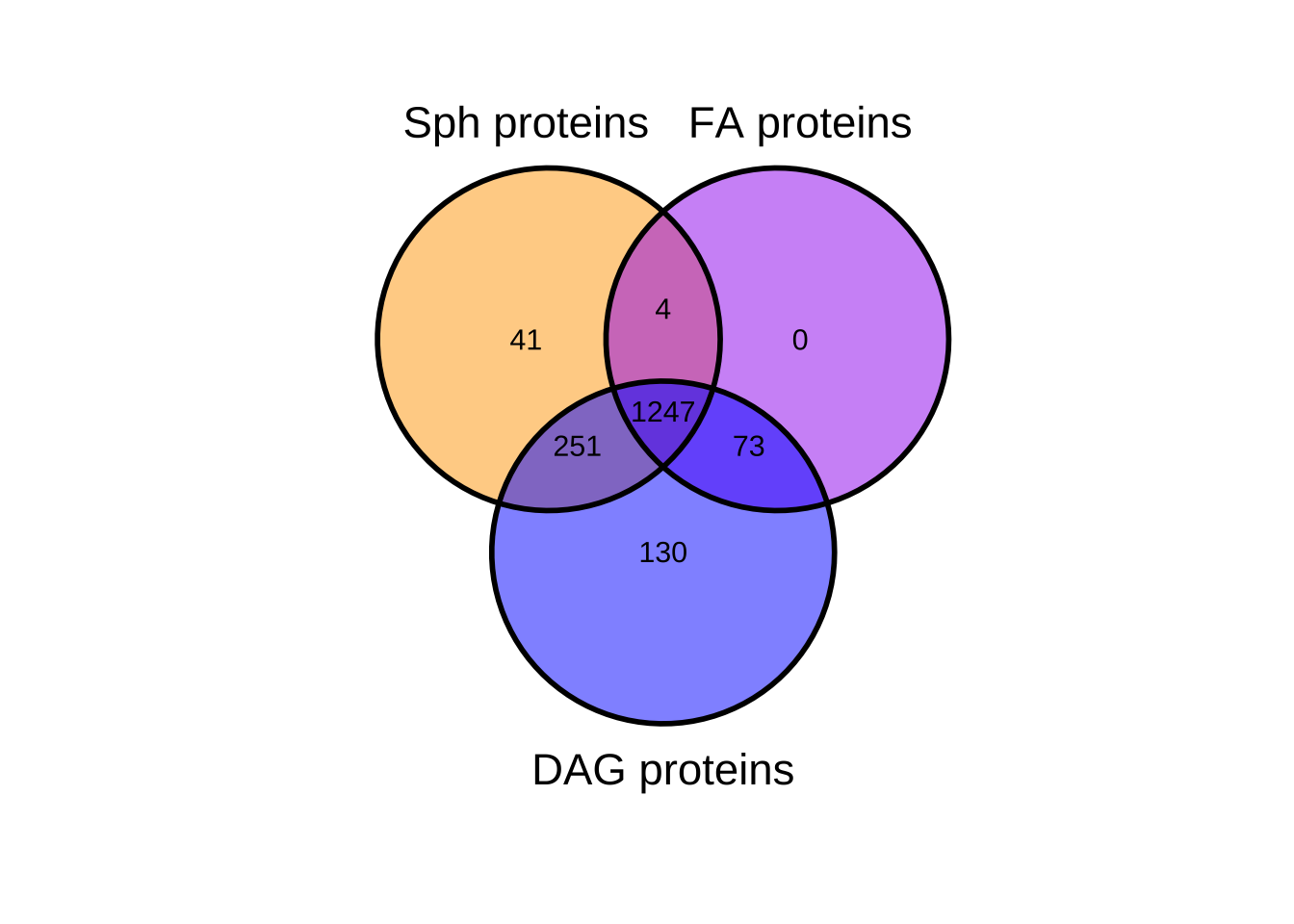
Venn Diagram
Gene Ontology Analysis
In beta: GO analysis still under development